In contrast to many venom labs around the world, we do not focus on specific groups of animals or toxins. Instead, we explore a wide diversity of organisms and biomolecules to gain holistic insights into the mechanistic basis of venom cocktails, their functions, evolution and translational potential. As a consequence, several projects are always running simultaneously within our group. A selected, but incomplete, list of projects is given below.
Venom Biotechnology
 Most neglected venomous animals belong to the smaller fraction of metazoan life forms, thus their venom yields limited amounts of toxins. Traditional approaches for Biodiscovery are largely pharmacology-driven, which means that venoms were subjected into chromatographic platforms, fractionated and tested. However this approach is rather sample-expensive and most neglected venomous organisms do not deliver required venom yields. Therefore, I apply biotechnological approaches to produce toxins identified in my other projects as an alternative. I utilize chemical synthesis, prokaryote plus eukaryote cell-free systems and recombinant expression strategies in E.coli via different strategies to produce otherwise untestable toxins in large quantities. Moreover, I apply bioengineering approaches to design artificial toxins, either as pharmacologically enhanced derivatives or for structure-function studies, from natural templates and express them alongside.
Most neglected venomous animals belong to the smaller fraction of metazoan life forms, thus their venom yields limited amounts of toxins. Traditional approaches for Biodiscovery are largely pharmacology-driven, which means that venoms were subjected into chromatographic platforms, fractionated and tested. However this approach is rather sample-expensive and most neglected venomous organisms do not deliver required venom yields. Therefore, I apply biotechnological approaches to produce toxins identified in my other projects as an alternative. I utilize chemical synthesis, prokaryote plus eukaryote cell-free systems and recombinant expression strategies in E.coli via different strategies to produce otherwise untestable toxins in large quantities. Moreover, I apply bioengineering approaches to design artificial toxins, either as pharmacologically enhanced derivatives or for structure-function studies, from natural templates and express them alongside.
Arthropod Venomics
 Arthropods are the most diverse group of terrestrial animals and several members evolved potent venom systems for prey subjugation and defence. However, only a marginal fraction of venoms throughout arthropods has been studied so far. In this project I aim to illuminate the venom systems of such arthropods, in particular of Central European spiders, via venom proteomics, transcriptomics and functional morphology. I am interested in the venom compositions, biological functions and the ecological roles of included molecules. Foremost I aim to identify novel venom components that may represent promising leads for subsequent modification into a therapeutic. I use insights from this project, in tandem with those from my more evolutionary projects to optimise the bioprospecting pipeline and to derive novel concepts on the idea of “promising taxa for Biodiscovery”.
Arthropods are the most diverse group of terrestrial animals and several members evolved potent venom systems for prey subjugation and defence. However, only a marginal fraction of venoms throughout arthropods has been studied so far. In this project I aim to illuminate the venom systems of such arthropods, in particular of Central European spiders, via venom proteomics, transcriptomics and functional morphology. I am interested in the venom compositions, biological functions and the ecological roles of included molecules. Foremost I aim to identify novel venom components that may represent promising leads for subsequent modification into a therapeutic. I use insights from this project, in tandem with those from my more evolutionary projects to optimise the bioprospecting pipeline and to derive novel concepts on the idea of “promising taxa for Biodiscovery”.
 Traditionally snakes and helodermatid lizards have been seen as the only venomous reptiles. However recent studies demonstrated that a venom system is ancestral to Ophidia and Anguimorpha, thus suggesting that more lizards evolved venom systems as previously acknowledged. In particular monitor lizards (Varanidae), including the enigmatig Komodo dragon (Varanus komodoensis) are nowadays often considered as venomous. In this collaborative project, my colleagues and I aim to disentangle putative venom systems of unstudied lizards, in particular of Varanidae and their kin. By combining venom proteomics, genomics, phylogenetic and functional morphology we want to resolve open questions on the evolutionary history of these lizards, deduce the functionality and composition of venom systems and, most importantly, try to understand the genomic processes that produce venom components in lizards.
Traditionally snakes and helodermatid lizards have been seen as the only venomous reptiles. However recent studies demonstrated that a venom system is ancestral to Ophidia and Anguimorpha, thus suggesting that more lizards evolved venom systems as previously acknowledged. In particular monitor lizards (Varanidae), including the enigmatig Komodo dragon (Varanus komodoensis) are nowadays often considered as venomous. In this collaborative project, my colleagues and I aim to disentangle putative venom systems of unstudied lizards, in particular of Varanidae and their kin. By combining venom proteomics, genomics, phylogenetic and functional morphology we want to resolve open questions on the evolutionary history of these lizards, deduce the functionality and composition of venom systems and, most importantly, try to understand the genomic processes that produce venom components in lizards. Spiders of the family Theraphosidae, commonly known as tarantulas, are the largest and probably most prominent spiders on earth. With ca. 1000 species they are the most diverse group of Mygalomorphae. That said, their systematics remain under critical discussion and the relationships within the family have not been disentangled. Moreover, a large fraction of tarantulas is frequently taxonomically revised and many taxa are relatively unstable. These effects are artefacts from the prevalent use of morphological characters in tarantula systematics. In this project, I aim to apply molecular Phylogenetics and Phylogenomics approaches to study tarantulas from a genetic perspective and to disentangle the relationships between major lineages within the family. I aim to develop novel wetlab protocols specific for tarantulas and to establish molecular technologies for theraphosid alpha-taxonomy. In combination with ethological data, Venomics and functional morphology I try to utilise the gained evolutionary insights on Theraphosidae to explore the origin and complexity of venom systems and other defences (urticating setae and stridulation) and to set these into ecological context.
Spiders of the family Theraphosidae, commonly known as tarantulas, are the largest and probably most prominent spiders on earth. With ca. 1000 species they are the most diverse group of Mygalomorphae. That said, their systematics remain under critical discussion and the relationships within the family have not been disentangled. Moreover, a large fraction of tarantulas is frequently taxonomically revised and many taxa are relatively unstable. These effects are artefacts from the prevalent use of morphological characters in tarantula systematics. In this project, I aim to apply molecular Phylogenetics and Phylogenomics approaches to study tarantulas from a genetic perspective and to disentangle the relationships between major lineages within the family. I aim to develop novel wetlab protocols specific for tarantulas and to establish molecular technologies for theraphosid alpha-taxonomy. In combination with ethological data, Venomics and functional morphology I try to utilise the gained evolutionary insights on Theraphosidae to explore the origin and complexity of venom systems and other defences (urticating setae and stridulation) and to set these into ecological context. Amphibians produce chemically complex defensive secretions, coined as skin poisons, in order to resist microbial colonisation and predation. A variety of powerful biomolecules is present in these including neurotoxic alkaloids, cardiotoxic steroids or haemolytic peptides – thus suggesting that amphibian poisons may harbour potent bioressources. It has recently been established that these skin poisons yield a subset of beneficial microbial-symbionts forming the “mucosome”, a largely unexplored micro-ecosystem in which toxins and bacteria are closely connected. In this project, I aim to explore the chemical composition of amphibian skin poisons, the role of bacteria within them and the ecological context in which these poisons work. I am interested in elucidating the potential of amphibian poisons plus their microbial symbionts to deliver novel pharmacologically active drugs and antibiotics. The principal, yet not exclusive, model group of this project are true salamanders of the genus Salamandra. A particular interest of mine is the discovery of novel derivatives of known toxin classes and to establish the interplay of toxicity and aposematic warning colorations.aaa
Amphibians produce chemically complex defensive secretions, coined as skin poisons, in order to resist microbial colonisation and predation. A variety of powerful biomolecules is present in these including neurotoxic alkaloids, cardiotoxic steroids or haemolytic peptides – thus suggesting that amphibian poisons may harbour potent bioressources. It has recently been established that these skin poisons yield a subset of beneficial microbial-symbionts forming the “mucosome”, a largely unexplored micro-ecosystem in which toxins and bacteria are closely connected. In this project, I aim to explore the chemical composition of amphibian skin poisons, the role of bacteria within them and the ecological context in which these poisons work. I am interested in elucidating the potential of amphibian poisons plus their microbial symbionts to deliver novel pharmacologically active drugs and antibiotics. The principal, yet not exclusive, model group of this project are true salamanders of the genus Salamandra. A particular interest of mine is the discovery of novel derivatives of known toxin classes and to establish the interplay of toxicity and aposematic warning colorations.aaa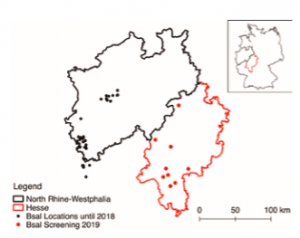 Amphibians are the most threatened vertebrates on earth and, in particular, emerging infectious diseases have recently imperilled amphibian populations globally. Among such, the “salamander eating fungus” Batrachochytrium salamandrivorans became of great concern for European species. It causes the lethal salamander plague in urodelans and especially members of Salamandra are susceptible to this disease. While salamander populations already collapsed in The Netherlands and Belgium, a current hotspot of Bsal is located in Germanies Eifel area in Northrine-westphalia. The federal state of Hesse harbours most of Germanies Salamandra populations and is located in close proximity to the infected area. Therefore, the government of Hesse has initiated a taskforce of conservationists and other scientists that aims to develop preventive measures for potential Bsal outbreaks in the future. In this taskforce I am educating citizen scientists in proper animal handling techniques, disinfection protocols and participate in various Bsal screenings of wild and captive specimen.
Amphibians are the most threatened vertebrates on earth and, in particular, emerging infectious diseases have recently imperilled amphibian populations globally. Among such, the “salamander eating fungus” Batrachochytrium salamandrivorans became of great concern for European species. It causes the lethal salamander plague in urodelans and especially members of Salamandra are susceptible to this disease. While salamander populations already collapsed in The Netherlands and Belgium, a current hotspot of Bsal is located in Germanies Eifel area in Northrine-westphalia. The federal state of Hesse harbours most of Germanies Salamandra populations and is located in close proximity to the infected area. Therefore, the government of Hesse has initiated a taskforce of conservationists and other scientists that aims to develop preventive measures for potential Bsal outbreaks in the future. In this taskforce I am educating citizen scientists in proper animal handling techniques, disinfection protocols and participate in various Bsal screenings of wild and captive specimen.